Kelvin waves
This tutorial describes the simulation of Kelvin waves propagating along nearly-straight and infinite vortex lines.
Here we will:
learn how to define infinite but unclosed filaments;
look at diagnostics such as the energy over time;
perform spatial and temporal Fourier analysis to detect relevant wavenumbers and frequencies associated to Kelvin waves.
It is recommended to first follow the vortex ring tutorial before following this tutorial.
Physical configuration
The idea of this tutorial is to study the time evolution of an infinite straight line slightly modified by a sinusoidal perturbation.
We will consider such a vortex line in a cubic periodic domain of size
Such an infinite line
for
The analytical prediction is that, over time, a small perturbation should rotate around the vortex in the direction opposite to its circulation. Its frequency is given by (see e.g. Schwarz (1985)):
where
Defining an unclosed infinite curve
Following the vortex ring tutorial, one may want to define such a line as follows:
using VortexPasta
using VortexPasta.Filaments
using VortexPasta.Filaments: Vec3
N = 64 # number of discretisation points per line
m = 2 # perturbation mode
L = 2π # domain period
x⃗₀ = Vec3(L/4, L/4, L/2) # line "origin"
ϵ = 0.01
ts = range(0, 1; length = N + 1)[1:N] # important: we exclude the endpoint (t = 1)
points = [x⃗₀ + Vec3(ϵ * L * sinpi(2m * t), 0, L * (t - 1/2)) for t ∈ ts]
f = Filaments.init(ClosedFilament, points, QuinticSplineMethod())Let's look at the result:
using GLMakie
# Give a colour to a filament based on its local orientation wrt Z.
function filament_colour(f::AbstractFilament, refinement)
cs = Float32[]
ζs = range(0, 1; length = refinement + 1)[1:refinement] # for interpolation
for seg ∈ segments(f), ζ ∈ ζs
colour = seg(ζ, UnitTangent())[3] # in [-1, 1]
push!(cs, colour)
end
let seg = last(segments(f)) # "close" the curve
colour = seg(1.0, UnitTangent())[3]
push!(cs, colour)
end
cs
end
# Plot a list of filaments
function plot_filaments(fs::AbstractVector)
fig = Figure()
ax = Axis3(fig[1, 1]; aspect = :data)
ticks = range(0, 2π; step = π/2)
tickformat(xs) = map(x -> string(x/π, "π"), xs)
ax.xticks = ax.yticks = ax.zticks = ticks
ax.xtickformat = ax.ytickformat = ax.ztickformat = tickformat
hidespines!(ax)
wireframe!(ax, Rect(0, 0, 0, L, L, L); color = (:black, 0.5), linewidth = 0.2)
for f ∈ fs
refinement = 4
color = filament_colour(f, refinement)
plot!(
ax, f;
refinement, color, colormap = :RdBu_9, colorrange = (-1, 1), markersize = 4,
)
end
fig
end
# Plot a single filament
plot_filaments(f::AbstractFilament) = plot_filaments([f])
plot_filaments(f)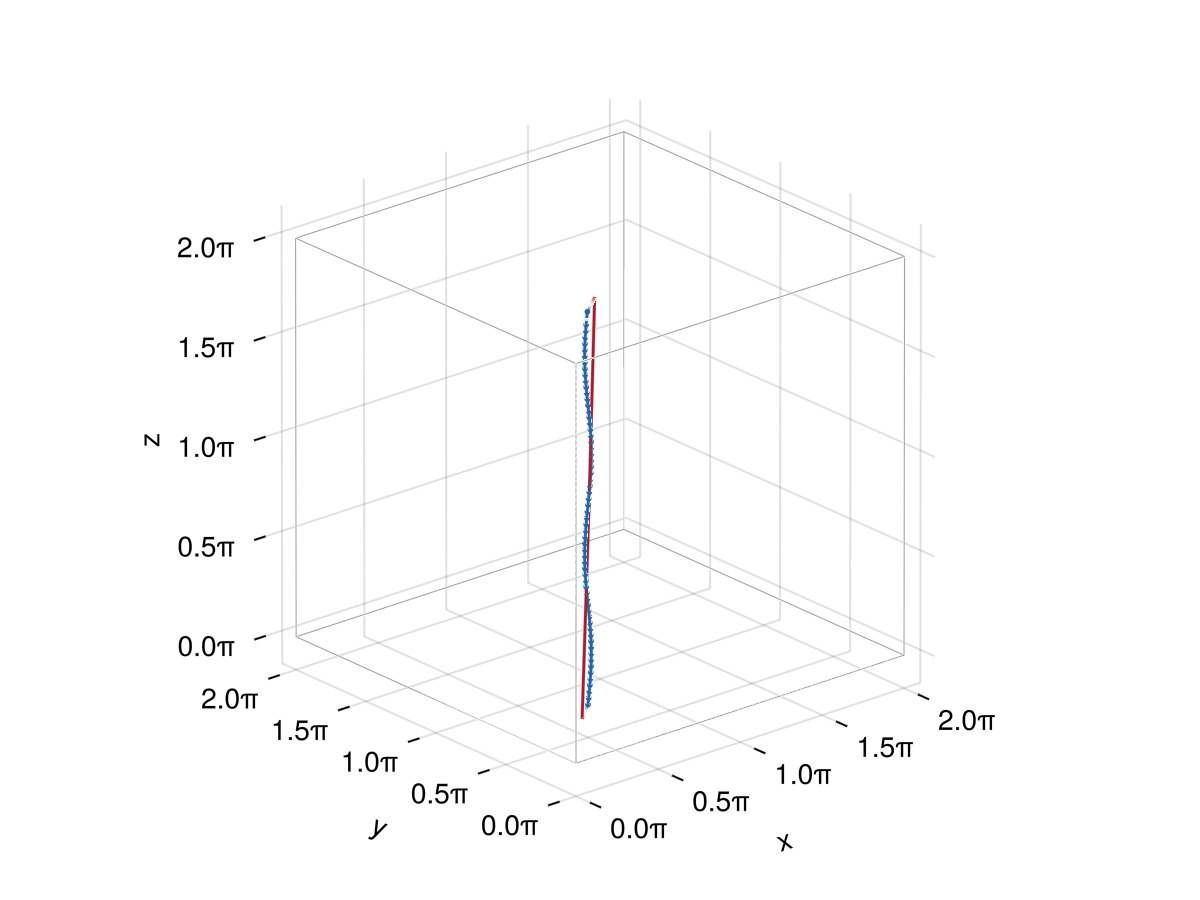
Things look almost as expected except for the fact that the line tries to close itself when it reaches the end. To avoid this, one needs to explicitly give Filaments.init an end-to-end vector via the offset keyword argument. In our case the end-to-end vector is
f = Filaments.init(ClosedFilament, points, QuinticSplineMethod(); offset = (0, 0, 2π))
plot_filaments(f)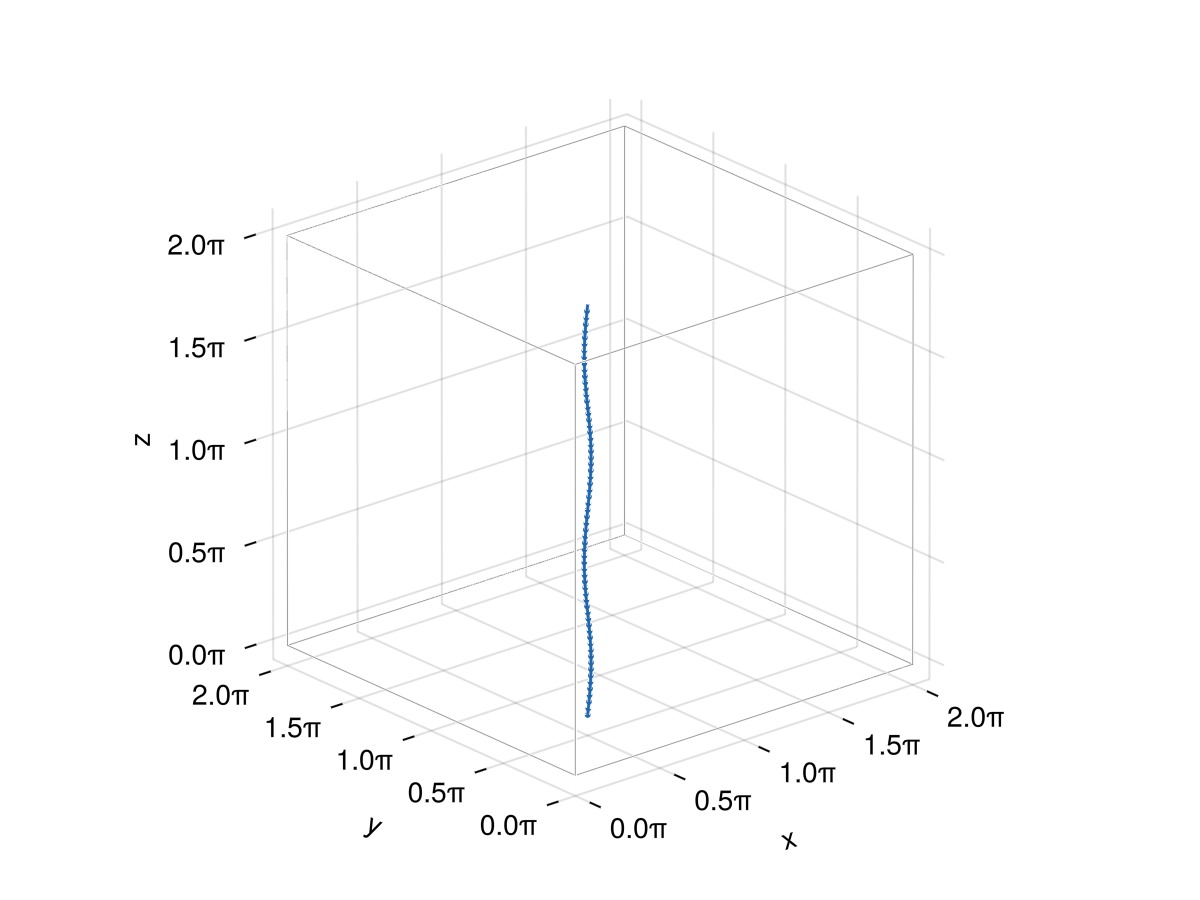
Now everything looks fine! Note that the end-to-end vector corresponds to the separation between a node f[i] and the node f[i + N]. For example:
@show f[end + 1] - f[begin]
@show Vec3(0, 0, 2π)f[end + 1] - f[begin] = [0.0, 0.0, 6.283185307179586]
Vec3(0, 0, 2π) = [0.0, 0.0, 6.283185307179586]End-to-end vector
The end-to-end vector must be an integer multiple of the domain period, which in this case is
Defining a curve from parametric function
The Filaments.init function actually allows to define a curve directly from its (continuous) parametric function. In this case one doesn't need to care about end-to-end vectors and "offsets", since these are usually encoded in the parametrisation.
For example, for the curve above we would define the function:
fcurve(t) = x⃗₀ + Vec3(
ϵ * L * sinpi(2 * m * t),
0,
(t - 0.5) * L,
)fcurve (generic function with 1 method)The function will be evaluated over the interval
a closed curve with period
an unclosed periodic curve which crosses the domain after a period
Here we are in the second case, and the function above indeed satisfies this condition.
Now we just pass the function to Filaments.init:
f_alt = Filaments.init(fcurve, ClosedFilament, N, QuinticSplineMethod())Note that this generates a filament which is practically identical to the previous one (just with a shift in the node positions, not really visible here):
plot_filaments([f, f_alt])
Using predefined curves
There is another convenient way of defining such curves, using the VortexPasta.PredefinedCurves module which provides definitions of parametric functions for commonly-used curves. As we will see in the next section, this is particularly convenient when we want to create multiple vortices which share the same geometry, but which have for instance different orientations or different spatial locations in the domain.
Here we want to use the PeriodicLine definitions, which allow one to pass arbitrary functions as perturbations. Note that curve definitions in PredefinedCurves are normalised. In particular, the period of PeriodicLine is 1, and the perturbation that we give it will be in terms of this unit period.
using VortexPasta.PredefinedCurves: PeriodicLine, define_curve
x_perturb(t) = ϵ * sinpi(2m * t) # perturbation along x (takes t ∈ [0, 1])
p = PeriodicLine(x = x_perturb) # this represents a line with period 1 along zWe now want to "convert" this line to a parametric function which can be then evaluated to generate points. This is done using the define_curve function, which allows in particular to rescale the curve (we want a period of
S = define_curve(p; scale = L, translate = x⃗₀)
@show S(0.0) S(0.5) S(1.0)S(0.0) = [1.5707963267948966, 1.5707963267948966, 0.0]
S(0.5) = [1.5707963267948966, 1.5707963267948966, 3.141592653589793]
S(1.0) = [1.5707963267948966, 1.5707963267948966, 6.283185307179586]As we can see, S is a function which can be evaluated at any value of S is identical to the fcurve function we defined above. We can now pass this function to Filaments.init to generate a filament:
f = Filaments.init(S, ClosedFilament, N, QuinticSplineMethod())
plot_filaments(f)
Ensuring periodicity of the velocity
For now we have initialised one infinite unclosed filament. One needs to be careful when working with unclosed filaments in periodic domains. Indeed, a single straight vortex filament in a periodic domain generates a non-zero circulation along the domain boundaries (or equivalently, a non-zero mean vorticity), which violates the periodicity condition. (One may compensate this by considering a uniform background vorticity field with opposite circulation, but this is outside of the scope of the tutorial.)
The mean vorticity in the periodic domain is given by
where
integrate(f_alt, GaussLegendre(4)) do ff, i, ζ
ff(i, ζ, Derivative(1))
end3-element SVector{3, Float64} with indices SOneTo(3):
-6.765421556309548e-17
-1.333667967889273e-29
6.283185307179587This means that, for each vortex oriented in the
In practice, to make sure that the total circulation is zero and to stabilise the system, we want to have four vortices such that their coordinates respect mirror symmetry with respect to the planes
Let's now create these four vortices:
funcs = [
# "Positive" vortices
define_curve(p; scale = (+L, +L, +L), translate = (0.25L, 0.25L, 0.5L)),
define_curve(p; scale = (-L, -L, +L), translate = (0.75L, 0.75L, 0.5L)), # mirror symmetry wrt x and y
# "Negative" vortices: we use the `orientation` keyword to flip their orientation.
define_curve(p; scale = (+L, -L, +L), translate = (0.25L, 0.75L, 0.5L), orientation = -1), # mirror symmetry wrt y
define_curve(p; scale = (-L, +L, +L), translate = (0.75L, 0.25L, 0.5L), orientation = -1), # mirror symmetry wrt x
]
fs = [Filaments.init(S, ClosedFilament, N, QuinticSplineMethod()) for S in funcs]
plot_filaments(fs)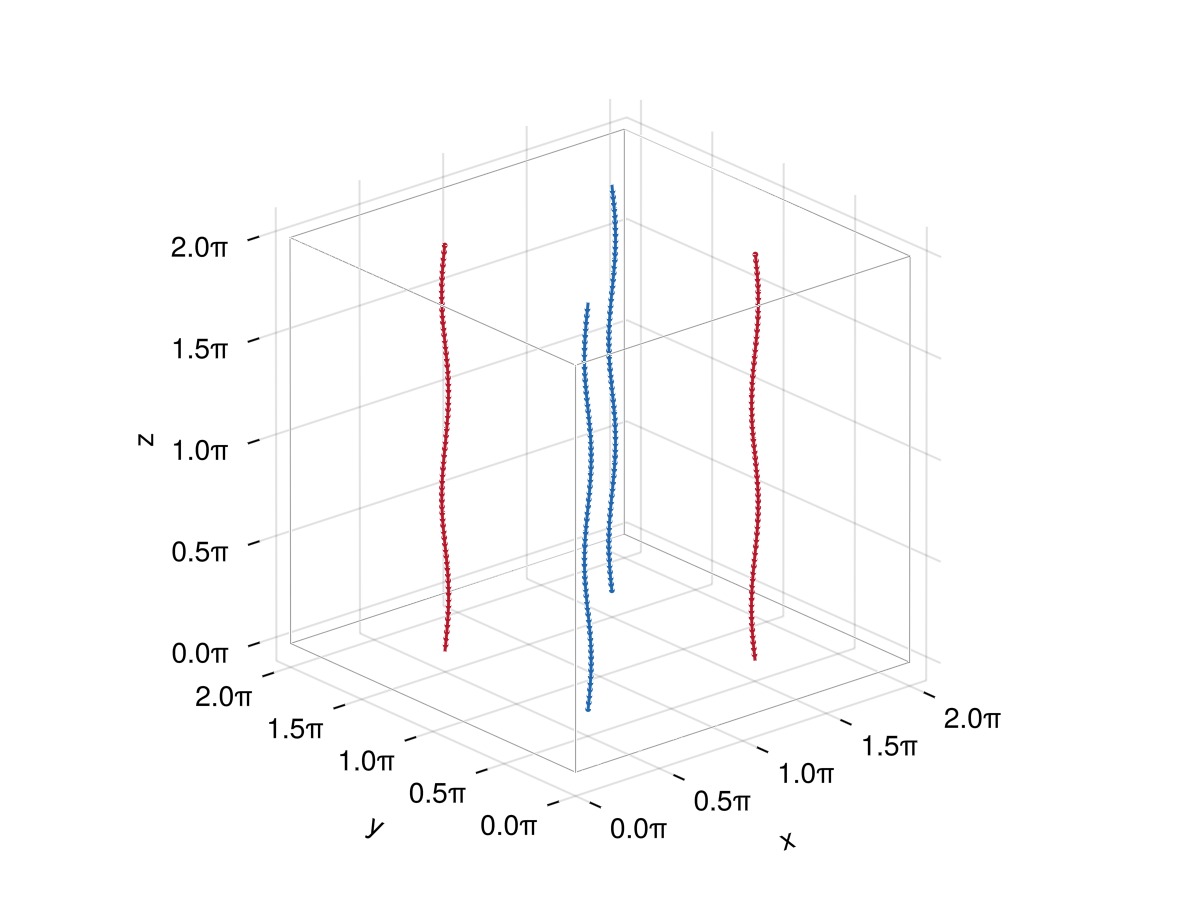
Here the colours represent the local orientation of the curve tangent with respect to the
# This computes the integral along each filament and sums the results.
sum(fs) do f
integrate(f, GaussLegendre(4)) do ff, i, ζ
f(i, ζ, Derivative(1))
end
end3-element SVector{3, Float64} with indices SOneTo(3):
-7.45931094670027e-17
-2.7739554174998235e-29
2.6645352591003757e-15Now we're ready to perform simulations.
Simulating Kelvin waves
As in the vortex ring tutorial, we use the Timestepping module to perform a temporal simulation of the configuration we just prepared.
Setting physical and numerical parameters
We start by setting the parameters for Biot–Savart computations:
using VortexPasta.BiotSavart
M = floor(Int, 32 * 2/3) # resolution of long-range grid
kmax = π * (M - 1) / L # maximum resolved wavenumber (Nyquist frequency) for long-range part
β = 3.5 # accuracy parameter
α = kmax / (2β) # Ewald splitting parameter
params = ParamsBiotSavart(;
Γ = 1.0, # vortex circulation
a = 1e-8, # vortex core size
Δ = 1/4, # vortex core parameter (1/4 for a constant vorticity distribution)
α = α, # Ewald splitting parameter
Ls = (L, L, L), # same domain size in all directions
Ns = (M, M, M), # same long-range resolution in all directions
rcut = β / α, # cut-off distance for short-range computations
quadrature = GaussLegendre(3), # quadrature for integrals over filament segments
backend_long = NonuniformFFTsBackend(), # this is the default
backend_short = CellListsBackend(2),
)ParamsBiotSavart{Float64} with:
- Physical parameters:
* Vortex circulation: Γ = 1.0
* Vortex core radius: a = 1.0e-8
* Vortex core parameter: Δ = 0.25
* Domain period: Ls = (6.283185307179586, 6.283185307179586, 6.283185307179586)
- Numerical parameters:
* Ewald splitting parameter: α = 1.4285714285714286 (σ = 1/α√2 = 0.49497474683058323)
* Quadrature rule: GaussLegendre{3}()
* Quadrature rule (alt.): GaussLegendre{3}() (used near singularities)
* Avoid explicit erf: true
* Short-range backend: CellListsBackend{2}(CPU(false); device = 1)
* Short-range cut-off: r_cut = 2.4499999999999997 (r_cut/L = 0.3899296105751435)
* Short-range cut-off coeff.: β_shortrange = 3.4999999999999996
* Local segment fraction: 1
* Short-range uses SIMD: true
* Long-range backend: NonuniformFFTsBackend(CPU(false); device = 1, m = HalfSupport(4), σ = 1.5)
* Long-range resolution: Ns = (21, 21, 21) (kmax = 10.0)
* Long-range cut-off coeff.: β_longrange = 3.5
* Long-range spherical truncation: falseWe would like to compute a few periods of Kelvin wave oscillations. For this, we first compute the expected Kelvin wave frequency and its associated period:
(; Γ, a, Δ,) = params # extract parameters needed for KW frequency
γ = MathConstants.eulergamma # Euler–Mascheroni constant
k = 2π * m / L
ω_kw = Γ * k^2 / (4 * π) * (
log(2 / (k * a)) - γ + 1/2 - Δ
)
T_kw = 2π / ω_kw # expected Kelvin wave period1.0909579075062508We create a VortexFilamentProblem to simulate a few Kelvin wave periods. To make things more interesting later when doing the temporal Fourier analysis of the results, we don't simulate an integer number of periods so that the results are not exactly time-periodic.
using VortexPasta.Timestepping
tspan = (0.0, 3.2 * T_kw)
prob = VortexFilamentProblem(fs, tspan, params)VortexFilamentProblem with fields:
├─ p: ParamsBiotSavart{Float64}(Γ = 1.0, a = 1.0e-8, Δ = 0.25, α = 1.4285714285714286, …)
├─ tspan: (0.0, 3.491065304020003) -- simulation timespan
└─ fs: 4-element VectorOfVectors -- vortex filaments at t = 0.0We now create a callback which will be used to store some data for further analysis. We will store the times and the position over time of a single filament node to be able to visualise and analyse the oscillations. Moreover, we will store the system energy to verify that energy is conserved over time (see VFM notes for detains on how it is computed). For computing the energy we use the kinetic_energy_from_streamfunction function from the Diagnostics module.
using VortexPasta.Diagnostics
# We use `const` for performance reasons, as these variables are defined in global scope
# and used within the callback. If we were inside a function, these would not be needed
# (and in fact Julia complains if we use `const` inside a function).
const times = Float64[]
const X_probe = Vec3{Float64}[] # will contain positions of a chosen node
const energy = Float64[]
# Save checkpoint file for visualisation every ≈ 0.1 * T_kw.
const t_checkpoint_step = 0.1 * T_kw
const t_checkpoint_next = Ref(tspan[1]) # save initial time, then every 0.1 * T_kw approx
function callback(iter)
(; nstep, t,) = iter
if nstep == 0 # make sure vectors are empty at the beginning of the simulation
empty!(times)
empty!(X_probe)
empty!(energy)
t_checkpoint_next[] = t
# Remove VTKHDF files from a previous run
for fname in readdir()
if match(r"^kelvin_waves_(\d+)\.vtkhdf$", fname) !== nothing
rm(fname)
end
end
end
push!(times, t)
s⃗ = iter.fs[1][3] # we choose a single node of a single filament
push!(X_probe, s⃗)
# Compute energy
E = Diagnostics.kinetic_energy(iter)
push!(energy, E)
# Save VTKHDF file for visualisation
if t ≥ t_checkpoint_next[]
save_checkpoint("kelvin_waves_$(nstep).vtkhdf", iter)
t_checkpoint_next[] += t_checkpoint_step # time at which the next file will be saved
end
nothing
endcallback (generic function with 1 method)Note that we have annotated the types of the variables times, X_probe and energy for performance reasons, since these are global variables which are used (and modified) from within the callback function. See here and here for details.
Choosing the timestep and the temporal scheme
In the vortex ring tutorial we have used the standard RK4 scheme. To capture the vortex evolution and avoid blow-up, this scheme requires the timestep
We first estimate the filament resolution using minimum_node_distance:
δ = minimum_node_distance(prob.fs) # should be close to L/N in our case0.0981747704246807Now we compute the Kelvin wave frequency associated to this distance:
kelvin_wave_period(λ; a, Δ, Γ) = 2 * λ^2 / Γ / (log(λ / (π * a)) + 1/2 - (Δ + MathConstants.γ))
dt_kw = kelvin_wave_period(δ; a, Δ, Γ)0.0013178102262909038Note that this time scale is very small compared to the period of the large-scale Kelvin waves:
T_kw / dt_kw827.8566107176529This means that we would need a relatively large simulation time to observe the evolution of large-scale Kelvin waves over multiple periods.
For the RK4 scheme, this time scale really seems to set the maximum allowed timestep limit. We can check that a simulation with RK4 using dt = dt_kw remains stable. In particular, energy stays constant in time after running a few iterations with this timestep:
iter = init(prob, RK4(); dt = dt_kw, callback)
for _ ∈ 1:40
step!(iter)
end
energy'1×41 adjoint(::Vector{Float64}) with eltype Float64:
0.159397 0.159397 0.159397 0.159397 … 0.159397 0.159397 0.159397However, using dt = 2 * dt_kw quickly leads to instability and energy blow-up:
iter = init(prob, RK4(); dt = 2 * dt_kw, callback)
for _ ∈ 1:40
step!(iter)
end
energy'1×41 adjoint(::Vector{Float64}) with eltype Float64:
0.159397 0.159397 0.159397 0.159397 … 0.324637 0.322037 0.323552This limit is basically set by the short-range Biot–Savart interactions and in particular the local term (see Desingularisation), which presents fast temporal variations. On the upside, this term is cheap to compute, which means that we can take advantage of a splitting scheme (such as Strang splitting) to accelerate computations.
The idea is to write the time evolution of a vortex point due to the Biot–Savart law as:
where the local term
Using a splitting method can make sense when it is easier or more convenient to separately solve the two equations:
In some applications, this is the case because one of the terms is linear or because one of the sub-equations can be solved analytically. This is not the case here, but this splitting is still convenient because it allows us to use different timesteps for each sub-equation. In particular, we can use a smaller timestep for the local (fast) term, which is the one that sets the timestep limit.
One of the most popular (and classical) splitting methods is Strang splitting, which is second order in time. In this method, a single simulation timestep (
Advance solution
. Advance solution
. Advance solution
.
In the following we use the Strang splitting scheme, which allows to use different explicit Runge–Kutta schemes for the "fast" and "slow" equations, and allows to set a smaller timestep to solve the former.
Concretely, we solve steps 1 and 3 using the standard RK4 scheme, while for step 2 the Midpoint scheme is used by default (but this can be changed, see Strang docs for details). Moreover, steps 1 and 3 are decomposed into
scheme = Strang(RK4(); nsubsteps = 16)
dt = 32 * dt_kw0.04216992724130892More generally, when using Strang splitting with the RK4 scheme for the fast dynamics, setting nsubsteps = M allows us to set the global timestep to dt = 2M * dt_kw. We could tune the number
Running the simulation
We now solve the full problem with this splitting scheme. Note that we use LocalTerm to identify the "fast" motions to the local (LIA) term of the Biot–Savart integrals. We could alternatively use ShortRangeTerm for a different interpretation of what represents the "fast" motions.
iter = init(prob, scheme; dt, callback, fast_term = LocalTerm())
reset_timer!(iter.to) # to get more accurate timings (removes most of the compilation time)
solve!(iter)
iter.to────────────────────────────────────────────────────────────────────────────────
Time Allocations
─────────────────────── ────────────────────────
Tot / % measured: 8.92s / 83.0% 634MiB / 70.9%
Section ncalls time %tot avg alloc %tot avg
────────────────────────────────────────────────────────────────────────────────
Update values at n... 10.9k 5.66s 76.5% 521μs 368MiB 81.9% 34.7KiB
Biot-Savart (LIA... 10.6k 3.77s 50.9% 355μs 264MiB 58.8% 25.5KiB
Add point charges 10.6k 1.84s 24.9% 173μs 45.2MiB 10.1% 4.36KiB
Short-range co... 10.6k 1.34s 18.1% 126μs 163MiB 36.2% 15.7KiB
Local term (... 10.6k 762ms 10.3% 71.8μs 22.7MiB 5.0% 2.19KiB
Copy point c... 10.6k 542ms 7.3% 51.0μs 127MiB 28.3% 12.3KiB
Synchronise GPU 10.6k 403μs 0.0% 37.9ns 0.00B 0.0% 0.00B
Copy output (d... 10.6k 357ms 4.8% 33.6μs 27.4MiB 6.1% 2.64KiB
Wait for async... 10.6k 14.3ms 0.2% 1.35μs 664KiB 0.1% 64.0B
Biot-Savart (exc... 166 903ms 12.2% 5.44ms 29.2MiB 6.5% 180KiB
Long-range com... 166 496ms 6.7% 2.99ms 16.6MiB 3.7% 102KiB
Vorticity to... 166 264ms 3.6% 1.59ms 7.42MiB 1.7% 45.8KiB
Interpolate ... 166 202ms 2.7% 1.22ms 5.99MiB 1.3% 36.9KiB
Velocity fie... 166 20.3ms 0.3% 122μs 573KiB 0.1% 3.45KiB
Copy point c... 166 8.78ms 0.1% 52.9μs 2.24MiB 0.5% 13.8KiB
Process poin... 166 187μs 0.0% 1.12μs 0.00B 0.0% 0.00B
Synchronise GPU 166 6.43μs 0.0% 38.8ns 0.00B 0.0% 0.00B
Short-range co... 166 161ms 2.2% 969μs 4.85MiB 1.1% 29.9KiB
Pair interac... 166 125ms 1.7% 752μs 1.05MiB 0.2% 6.47KiB
Remove self-... 166 21.3ms 0.3% 128μs 408KiB 0.1% 2.46KiB
Copy point c... 166 10.4ms 0.1% 62.4μs 2.49MiB 0.6% 15.4KiB
Process poin... 166 3.14ms 0.0% 18.9μs 622KiB 0.1% 3.75KiB
Synchronise GPU 166 6.38μs 0.0% 38.4ns 0.00B 0.0% 0.00B
Add point charges 166 27.3ms 0.4% 165μs 724KiB 0.2% 4.36KiB
Copy output (d... 332 12.8ms 0.2% 38.6μs 926KiB 0.2% 2.79KiB
Wait for async... 332 315μs 0.0% 949ns 20.8KiB 0.0% 64.0B
CPU-only opera... 166 13.9μs 0.0% 83.8ns 0.00B 0.0% 0.00B
Biot-Savart (full) 83 490ms 6.6% 5.91ms 17.0MiB 3.8% 210KiB
Long-range com... 83 368ms 5.0% 4.44ms 11.6MiB 2.6% 143KiB
Interpolate ... 166 210ms 2.8% 1.26ms 6.01MiB 1.3% 37.0KiB
Vorticity to... 83 132ms 1.8% 1.59ms 3.72MiB 0.8% 45.9KiB
Velocity fie... 83 12.2ms 0.2% 147μs 287KiB 0.1% 3.45KiB
Streamfuncti... 83 9.20ms 0.1% 111μs 290KiB 0.1% 3.49KiB
Copy point c... 83 4.45ms 0.1% 53.6μs 1.12MiB 0.2% 13.8KiB
Process poin... 83 114μs 0.0% 1.38μs 0.00B 0.0% 0.00B
Synchronise GPU 83 3.41μs 0.0% 41.0ns 0.00B 0.0% 0.00B
Short-range co... 83 94.6ms 1.3% 1.14ms 3.64MiB 0.8% 44.9KiB
Pair interac... 83 64.6ms 0.9% 778μs 550KiB 0.1% 6.63KiB
Remove self-... 83 11.1ms 0.1% 133μs 211KiB 0.0% 2.54KiB
Copy point c... 166 8.75ms 0.1% 52.7μs 2.24MiB 0.5% 13.8KiB
Local term (... 83 7.95ms 0.1% 95.7μs 187KiB 0.0% 2.25KiB
Process poin... 83 1.62ms 0.0% 19.5μs 311KiB 0.1% 3.75KiB
Synchronise GPU 83 3.38μs 0.0% 40.8ns 0.00B 0.0% 0.00B
Add point charges 83 15.6ms 0.2% 187μs 364KiB 0.1% 4.38KiB
Copy output (d... 166 9.04ms 0.1% 54.5μs 878KiB 0.2% 5.29KiB
Wait for async... 166 193μs 0.0% 1.16μs 10.4KiB 0.0% 64.0B
CPU-only opera... 83 93.5μs 0.0% 1.13μs 0.00B 0.0% 0.00B
Advect filaments 10.9k 1.61s 21.7% 148μs 70.9MiB 15.8% 6.68KiB
Callback 83 113ms 1.5% 1.36ms 8.38MiB 1.9% 103KiB
After advection 83 9.46ms 0.1% 114μs 335KiB 0.1% 4.04KiB
Fold and refine 83 8.93ms 0.1% 108μs 320KiB 0.1% 3.85KiB
Reconnect 83 207μs 0.0% 2.50μs 6.48KiB 0.0% 80.0B
Prepare interpolab... 83 9.08ms 0.1% 109μs 1.78MiB 0.4% 21.9KiB
Resize timesteppin... 83 176μs 0.0% 2.12μs 0.00B 0.0% 0.00B
────────────────────────────────────────────────────────────────────────────────We can check that energy is conserved:
energy'1×84 adjoint(::Vector{Float64}) with eltype Float64:
0.159397 0.159397 0.159397 0.159397 … 0.159397 0.159397 0.159397We see that the energy seems to take the same value at all times. We can verify this quantitatively by looking at its standard deviation (normalised by the mean energy), which is negligible:
using Statistics: mean, std
Emean = mean(energy)
Estd = std(energy)
Estd / Emean1.5787602922994016e-8We now plot the evolution of the
fig = Figure()
ax = Axis(fig[1, 1]; xlabel = L"t / T_{\text{KW}}", ylabel = "Position")
tnorm = times ./ T_kw # normalised time
xpos = [s⃗[1] for s⃗ in X_probe] # get all X positions over time
ypos = [s⃗[2] for s⃗ in X_probe] # get all Y positions over time
scatterlines!(ax, tnorm, xpos; marker = 'o', label = L"x(t)")
scatterlines!(ax, tnorm, ypos; marker = 'o', label = L"y(t)")
Legend(fig[1, 2], ax; orientation = :vertical, framevisible = false, padding = (0, 0, 0, 0), labelsize = 22, rowgap = 5)
fig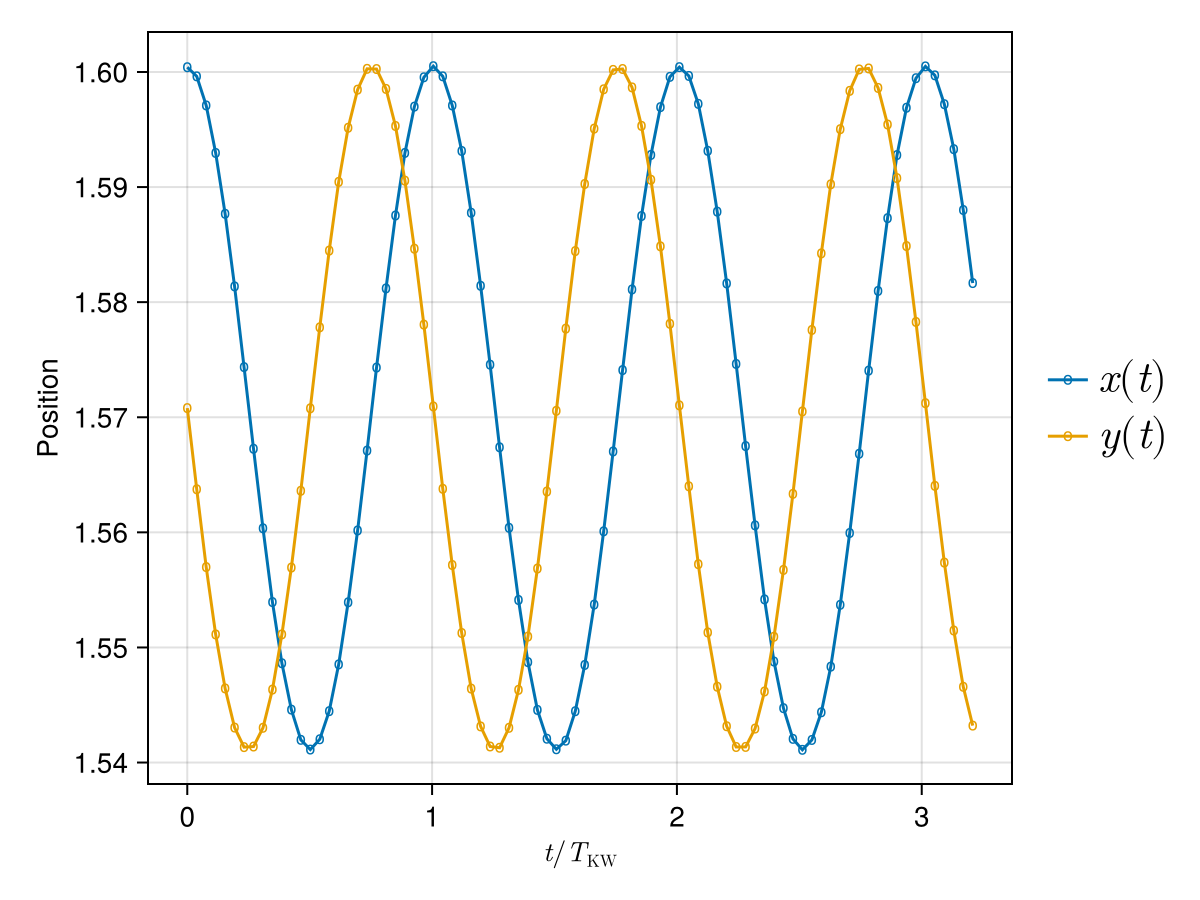
We see that the
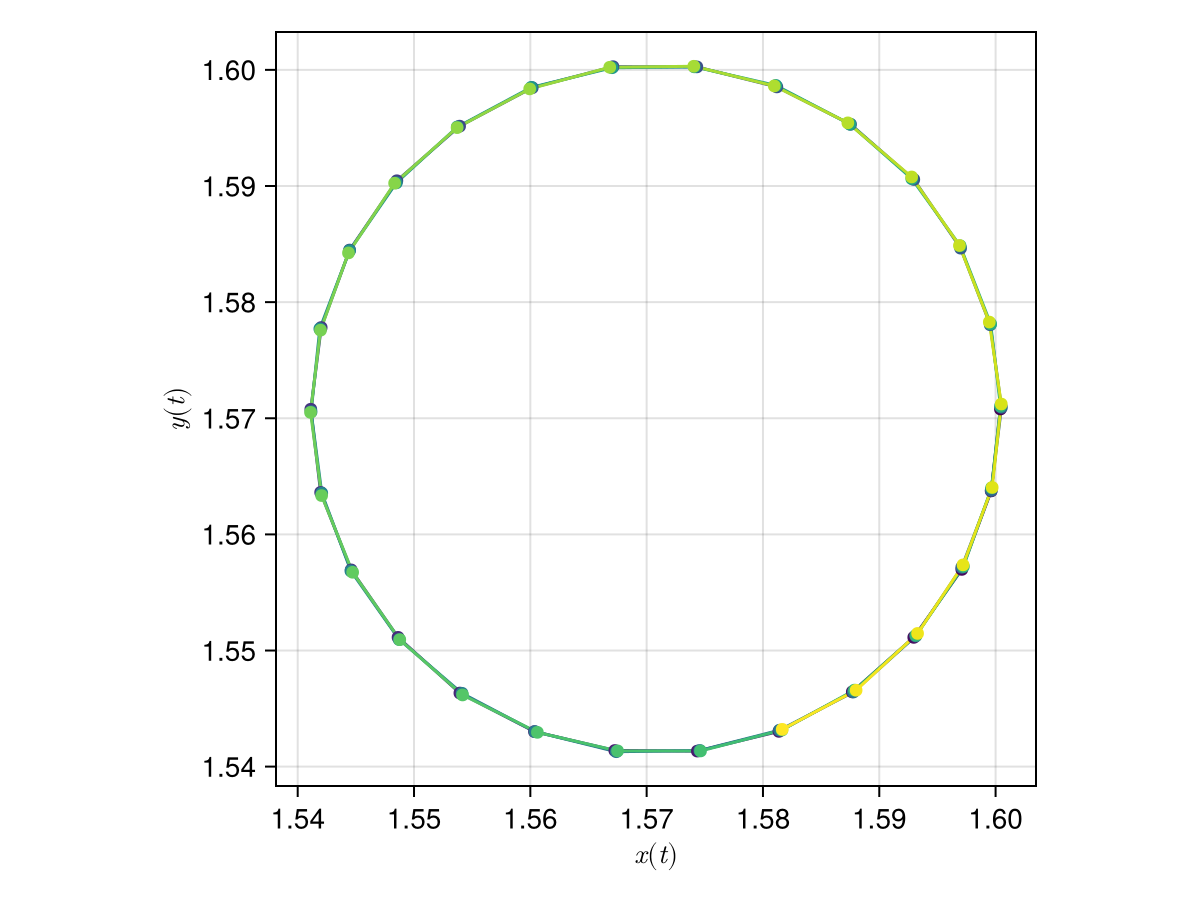
The oscillations above suggest circular trajectories, as we can check in the following figure:
scatterlines(
xpos, ypos;
color = tnorm,
axis = (aspect = DataAspect(), xlabel = L"x(t)", ylabel = L"y(t)"),
)
Measuring performance
The VortexPasta solver uses the TimerOutputs.jl package to estimate the time spent (and memory allocated) in different stages of the computation.
Accessing timing information is very simple, as it is all included in the to field of the VortexFilamentSolver:
iter.to──────────────────────────────────────────────────────────────────────────────────────────────────────
Time Allocations
─────────────────────── ────────────────────────
Tot / % measured: 17.2s / 43.0% 1.51GiB / 29.0%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────────────────────────────
Update values at nodes 10.9k 5.66s 76.5% 521μs 368MiB 81.9% 34.7KiB
Biot-Savart (LIA only) 10.6k 3.77s 50.9% 355μs 264MiB 58.8% 25.5KiB
Add point charges 10.6k 1.84s 24.9% 173μs 45.2MiB 10.1% 4.36KiB
Short-range component (async) 10.6k 1.34s 18.1% 126μs 163MiB 36.2% 15.7KiB
Local term (LIA) 10.6k 762ms 10.3% 71.8μs 22.7MiB 5.0% 2.19KiB
Copy point charges (host -> device) 10.6k 542ms 7.3% 51.0μs 127MiB 28.3% 12.3KiB
Synchronise GPU 10.6k 403μs 0.0% 37.9ns 0.00B 0.0% 0.00B
Copy output (device -> host) 10.6k 357ms 4.8% 33.6μs 27.4MiB 6.1% 2.64KiB
Wait for async task to finish 10.6k 14.3ms 0.2% 1.35μs 664KiB 0.1% 64.0B
Biot-Savart (excluding LIA) 166 903ms 12.2% 5.44ms 29.2MiB 6.5% 180KiB
Long-range component (async) 166 496ms 6.7% 2.99ms 16.6MiB 3.7% 102KiB
Vorticity to Fourier 166 264ms 3.6% 1.59ms 7.42MiB 1.7% 45.8KiB
Interpolate to physical 166 202ms 2.7% 1.22ms 5.99MiB 1.3% 36.9KiB
Velocity field (Fourier) 166 20.3ms 0.3% 122μs 573KiB 0.1% 3.45KiB
Copy point charges (host -> device) 166 8.78ms 0.1% 52.9μs 2.24MiB 0.5% 13.8KiB
Process point charges 166 187μs 0.0% 1.12μs 0.00B 0.0% 0.00B
Synchronise GPU 166 6.43μs 0.0% 38.8ns 0.00B 0.0% 0.00B
Short-range component (async) 166 161ms 2.2% 969μs 4.85MiB 1.1% 29.9KiB
Pair interactions 166 125ms 1.7% 752μs 1.05MiB 0.2% 6.47KiB
Remove self-interactions 166 21.3ms 0.3% 128μs 408KiB 0.1% 2.46KiB
Copy point charges (host -> device) 166 10.4ms 0.1% 62.4μs 2.49MiB 0.6% 15.4KiB
Process point charges 166 3.14ms 0.0% 18.9μs 622KiB 0.1% 3.75KiB
Synchronise GPU 166 6.38μs 0.0% 38.4ns 0.00B 0.0% 0.00B
Add point charges 166 27.3ms 0.4% 165μs 724KiB 0.2% 4.36KiB
Copy output (device -> host) 332 12.8ms 0.2% 38.6μs 926KiB 0.2% 2.79KiB
Wait for async task to finish 332 315μs 0.0% 949ns 20.8KiB 0.0% 64.0B
CPU-only operations (synchronous) 166 13.9μs 0.0% 83.8ns 0.00B 0.0% 0.00B
Biot-Savart (full) 83 490ms 6.6% 5.91ms 17.0MiB 3.8% 210KiB
Long-range component (async) 83 368ms 5.0% 4.44ms 11.6MiB 2.6% 143KiB
Interpolate to physical 166 210ms 2.8% 1.26ms 6.01MiB 1.3% 37.0KiB
Vorticity to Fourier 83 132ms 1.8% 1.59ms 3.72MiB 0.8% 45.9KiB
Velocity field (Fourier) 83 12.2ms 0.2% 147μs 287KiB 0.1% 3.45KiB
Streamfunction field (Fourier) 83 9.20ms 0.1% 111μs 290KiB 0.1% 3.49KiB
Copy point charges (host -> device) 83 4.45ms 0.1% 53.6μs 1.12MiB 0.2% 13.8KiB
Process point charges 83 114μs 0.0% 1.38μs 0.00B 0.0% 0.00B
Synchronise GPU 83 3.41μs 0.0% 41.0ns 0.00B 0.0% 0.00B
Short-range component (async) 83 94.6ms 1.3% 1.14ms 3.64MiB 0.8% 44.9KiB
Pair interactions 83 64.6ms 0.9% 778μs 550KiB 0.1% 6.63KiB
Remove self-interactions 83 11.1ms 0.1% 133μs 211KiB 0.0% 2.54KiB
Copy point charges (host -> device) 166 8.75ms 0.1% 52.7μs 2.24MiB 0.5% 13.8KiB
Local term (LIA) 83 7.95ms 0.1% 95.7μs 187KiB 0.0% 2.25KiB
Process point charges 83 1.62ms 0.0% 19.5μs 311KiB 0.1% 3.75KiB
Synchronise GPU 83 3.38μs 0.0% 40.8ns 0.00B 0.0% 0.00B
Add point charges 83 15.6ms 0.2% 187μs 364KiB 0.1% 4.38KiB
Copy output (device -> host) 166 9.04ms 0.1% 54.5μs 878KiB 0.2% 5.29KiB
Wait for async task to finish 166 193μs 0.0% 1.16μs 10.4KiB 0.0% 64.0B
CPU-only operations (synchronous) 83 93.5μs 0.0% 1.13μs 0.00B 0.0% 0.00B
Advect filaments 10.9k 1.61s 21.7% 148μs 70.9MiB 15.8% 6.68KiB
Callback 83 113ms 1.5% 1.36ms 8.38MiB 1.9% 103KiB
After advection 83 9.46ms 0.1% 114μs 335KiB 0.1% 4.04KiB
Fold and refine 83 8.93ms 0.1% 108μs 320KiB 0.1% 3.85KiB
Reconnect 83 207μs 0.0% 2.50μs 6.48KiB 0.0% 80.0B
Prepare interpolable fields 83 9.08ms 0.1% 109μs 1.78MiB 0.4% 21.9KiB
Resize timestepping cache 83 176μs 0.0% 2.12μs 0.00B 0.0% 0.00B
──────────────────────────────────────────────────────────────────────────────────────────────────────We can see that, in this particular case, the runtime is mostly dominated by the the LIA (local) term, which is computed much more often than the non-local interactions due to the use of a splitting scheme for the temporal evolution.
Fourier analysis
Spatial analysis
The idea is to identify the spatial fluctuations of a single vortex with respect to the unperturbed filament. For this, we first write the perturbations in complex representation as a function of the
We want to apply the FFT to these two functions. For this, we need all points of the vortex filament to be equispaced in
f = iter.fs[1] # vortex to analyse
zs = [s⃗.z for s⃗ in f] # y locations
N = length(zs)
zs_expected = range(zs[begin], zs[begin] + L; length = N + 1)[1:N] # equispaced locations
isapprox(zs, zs_expected; rtol = 1e-5) # check that z locations are approximately equispacedtrueNow that we have verified this, we define the complex function
xs = [s⃗.x for s⃗ in f] # x locations
ys = [s⃗.y for s⃗ in f] # y locations
ws = @. xs + im * ys
using FFTW: fft, fft!, fftfreq, fftshift
w_hat = fft(ws)
@. w_hat = w_hat / N # normalise FFT
@show w_hat[1] # the zero frequency gives the mean location
w_hat[1] ≈ π/2 + π/2 * im # we expect the mean location to be (π/2, π/2)trueThe associated wavenumbers are multiples of
Δz = L / N
@assert isapprox(Δz, zs[2] - zs[1]; rtol = 1e-4)
ks = fftfreq(N, 2π / Δz)
ks' # should be integers if L = 2π1×64 adjoint(::AbstractFFTs.Frequencies{Float64}) with eltype Float64:
0.0 1.0 2.0 3.0 4.0 5.0 6.0 … -6.0 -5.0 -4.0 -3.0 -2.0 -1.0Note that this includes positive and negative wavenumbers. More precisely, ks[2:N÷2] contains the positive wavenumbers, and ks[N÷2+1:end] contains the corresponding negative wavenumbers.
We now want to compute the wave action spectrum
function wave_action_spectrum(ks::AbstractVector, w_hat::AbstractVector)
@assert ks[2] == -ks[end] # contains positive and negative wavenumbers
@assert length(ks) == length(w_hat)
N = length(ks)
dk = ks[2] # this is the wavenumber increment
if iseven(N)
Nh = N ÷ 2
@assert ks[Nh + 1] ≈ -(ks[Nh] + dk) # wavenumbers change sign after index Nh
else
Nh = N ÷ 2 + 1
@assert ks[Nh + 1] == -ks[Nh] # wavenumbers change sign after index Nh
end
ks_pos = ks[2:Nh] # only positive wavenumbers
nk = similar(ks_pos)
for j ∈ eachindex(ks_pos)
local k = ks_pos[j]
i⁺ = 1 + j # index of coefficient corresponding to wavenumber +k
i⁻ = N + 1 - j # index of coefficient corresponding to wavenumber -k
@assert ks[i⁺] == -ks[i⁻] == k # verification
nk[j] = abs2(w_hat[i⁺]) + abs2(w_hat[i⁻])
end
ks_pos, nk
end
ks_pos, nk = wave_action_spectrum(ks, w_hat)
nk_normalised = nk ./ ((ϵ * L)^2 / 2)
sum(nk_normalised) # we expect the sum to be 10.999994396307008We can finally plot the final state:
fig = Figure()
ax = Axis(
fig[1, 1];
xscale = log10, yscale = log10, xlabel = L"k", ylabel = L"2 \, n(k) / (ϵ L)^2",
xlabelsize = 20, ylabelsize = 20,
xticks = LogTicks(0:4), xminorticksvisible = true, xminorticks = IntervalsBetween(9),
yminorticksvisible = true, yminorticks = exp10.(-40:10),
)
scatterlines!(ax, ks_pos, nk_normalised)
xlims!(ax, 0.8 * ks_pos[begin], nothing)
ylims!(ax, 1e-30, 1e1)
vlines!(ax, ks_pos[m]; linestyle = :dash, color = :orangered)
fig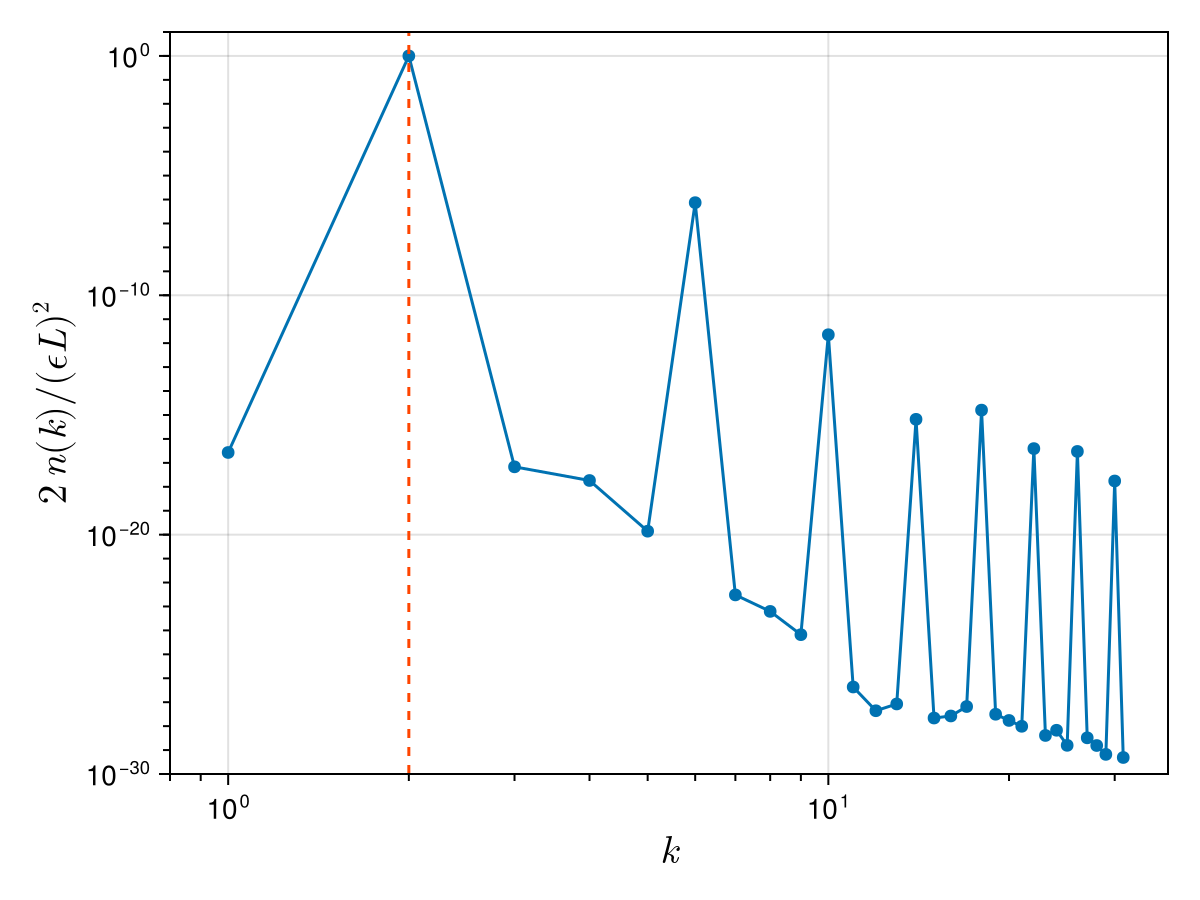
We see that the wave action spectrum is strongly peaked at the wavenumber
A kind of proof using LIA
One way to illustrate this analytically is using the local induction approximation (LIA). Let's consider the perturbed line
where
In our case we have
Using a Taylor expansion, one can easily show that the denominator introduces fluctuations over the odd harmonics
Noting that
In the tutorial, we initially perturbed the mode
We also see that the sum
The main conclusion is that, when we perturb a single Kelvin wave mode as we did here, that original mode is exactly preserved over time (except for negligible high-order effects).
Temporal analysis
We can do something similar to analyse the temporal oscillations of the filament. For example, we can take the same temporal data we analysed before, corresponding to the position of a single filament node:
xt = getindex.(X_probe, 1) # x positions of a single node over time
yt = getindex.(X_probe, 2) # y positions of a single node over time
zt = getindex.(X_probe, 3) # z positions of a single node over time
std(zt) / mean(zt) # ideally, the z positions shouldn't change over time1.6622057240119106e-5Similarly to before, we now write
inds_t = eachindex(times)[begin:end - 1] # don't consider the last time to make sure the timestep Δt is constant
w_equilibrium = 0.25L + im * 0.25L # equilibrium position of first vortex (w = x + iy)
wt = @views @. xt[inds_t] + im * yt[inds_t] - w_equilibrium # we subtract the equilibrium position
Nt = length(wt) # number of time snapshots
Δt = times[2] - times[1] # timestep
@assert times[begin:end-1] ≈ range(times[begin], times[end-1]; length = Nt) # check that times are equispaced
w_hat = fft(wt)
@. w_hat = w_hat / Nt # normalise FFT
ωs = fftfreq(Nt, 2π / Δt)
ωs_pos, nω = wave_action_spectrum(ωs, w_hat)
ωs_normalised = ωs_pos ./ ω_kw # normalise by expected KW frequency
fig = Figure()
ax = Axis(
fig[1, 1];
xscale = log10, yscale = log10, xlabel = L"ω / ω_{\text{kw}}", ylabel = L"n(ω)",
xlabelsize = 20, ylabelsize = 20,
xticks = LogTicks(0:4), xminorticksvisible = true, xminorticks = IntervalsBetween(9),
yticks = LogTicks(-20:10), yminorticksvisible = true, yminorticks = IntervalsBetween(9),
)
scatterlines!(ax, ωs_normalised, nω; label = "Original signal")
xlims!(ax, 0.8 * ωs_normalised[begin], 1.2 * ωs_normalised[end])
vlines!(ax, 1.0; linestyle = :dash, color = :orangered)
fig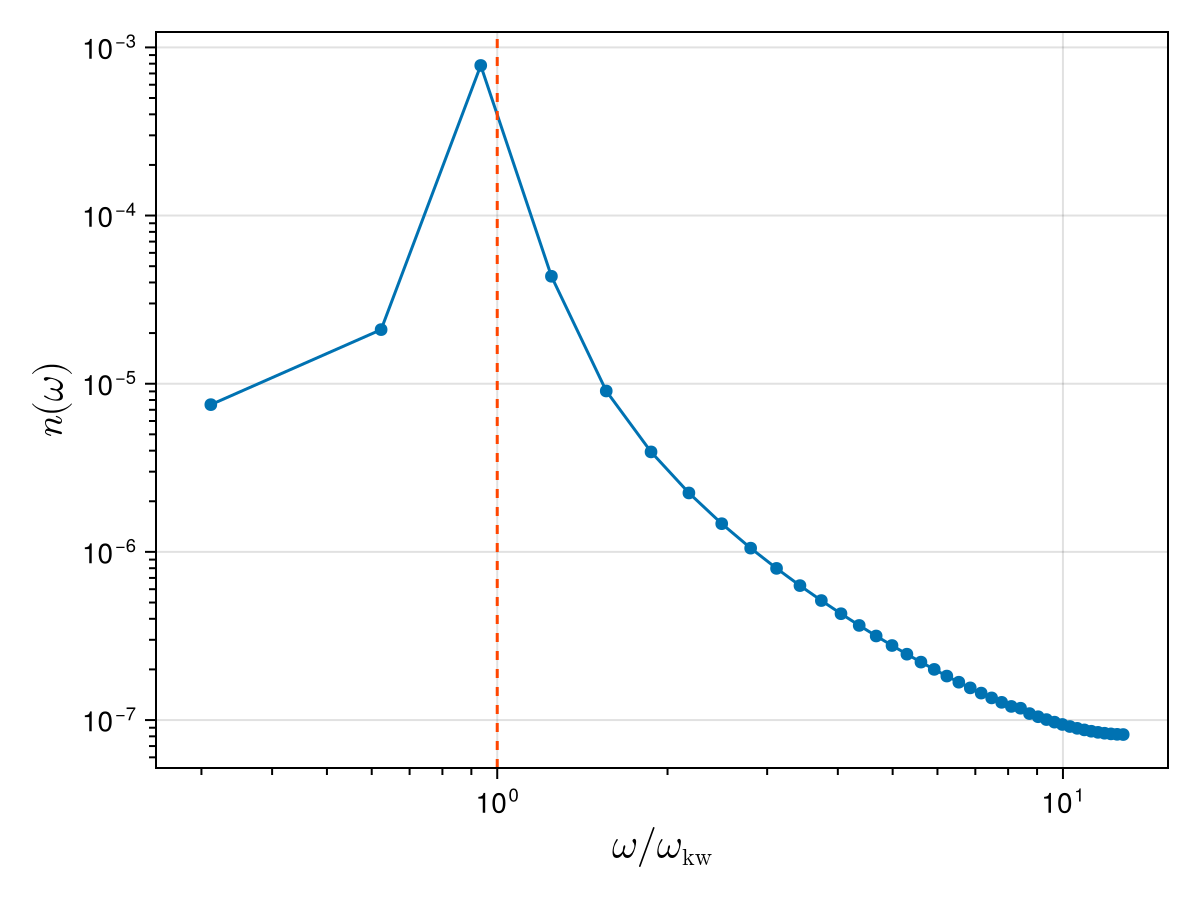
We see that the temporal spectrum is strongly peaked near the analytical Kelvin wave frequency (dashed vertical line). However, since the trajectory is not perfectly periodic in time (the signal is discontinuous when going from the final time to the initial time), other frequencies are also present in the spectrum (this is known as spectral leakage).
To reduce the effect of spectral leakage, the usual solution is to apply a window function to the original signal to make it periodic. There are many examples of window functions which are commonly used in signal processing.
Here we use the DSP.jl package which includes many definitions of window functions. Note that we first need to subtract the mean value from our input signal before multiplying it by the window function. Below we compare the previous temporal spectrum with the one obtained after applying the Hann window:
using DSP: DSP
wt_mean = mean(wt)
window = DSP.Windows.hanning(Nt)
wt_windowed = @. (wt - wt_mean) * window
w_hat = fft(wt_windowed)
@. w_hat = w_hat / Nt # normalise FFT
_, nω_windowed = wave_action_spectrum(ωs, w_hat)
scatterlines!(ax, ωs_normalised, nω_windowed; label = "Windowed signal")
Legend(fig[0, 1], ax; orientation = :horizontal, framevisible = false, colgap = 32, patchsize = (40, 10))
rowgap!(fig[:, 1].layout, 6) # reduce gap between plot and legend (default gap is 18)
fig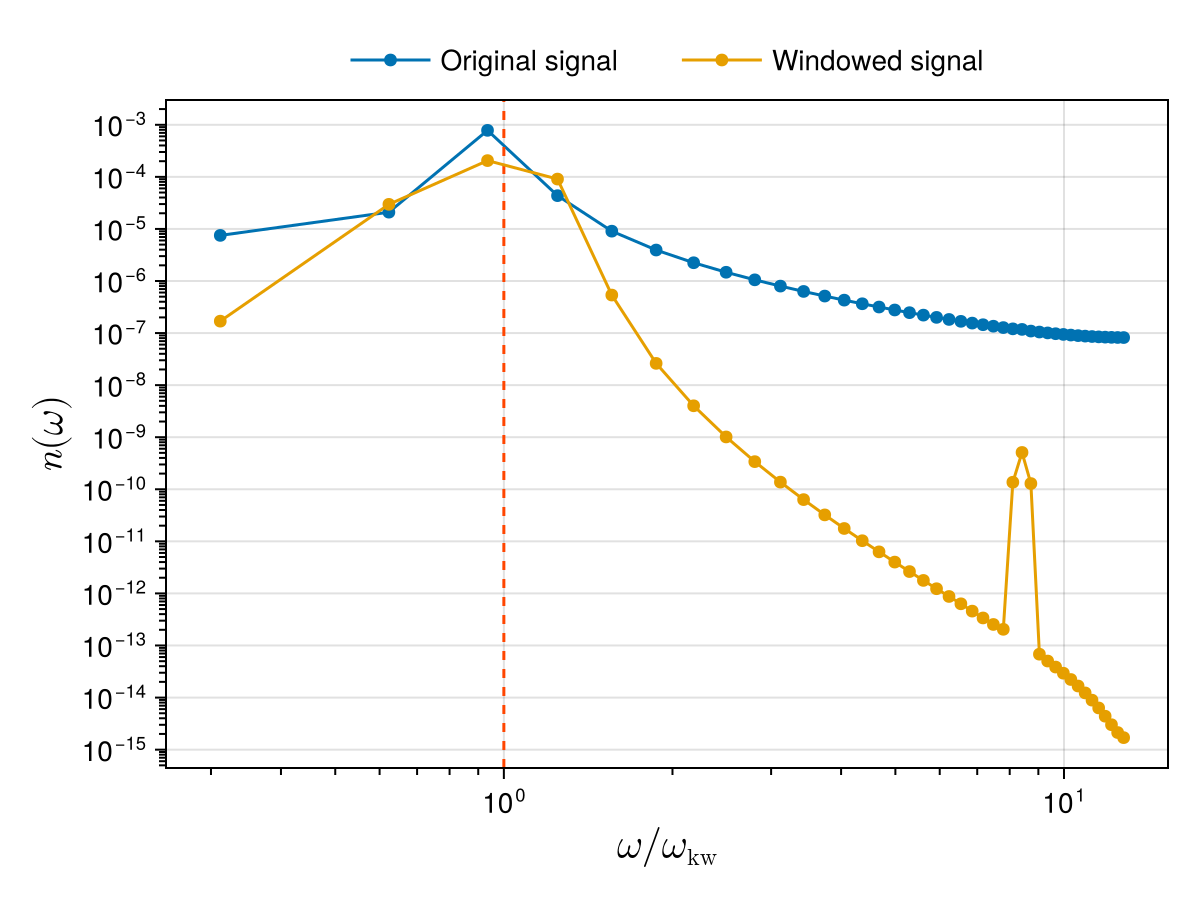
The new spectrum is still peaked near the expected frequency, while artificial modes far from this frequency are strongly damped compared to the original spectrum. Note however that windowing tends to smoothen the spectrum around the analytical Kelvin wave frequency.
Dispersion relation
To verify the dispersion relation of Kelvin waves, we perform a new simulation where we initially excite a large range of wavenumbers
Creating the initial condition
We start by defining one straight filament:
p = PeriodicLine() # no perturbation, we will perturb it afterwards
w_equilibrium = 0.25L + im * 0.25L # equilibrium position of vortex (w = x + iy)
S = define_curve(p; scale = (+L, +L, +L), translate = (0.25L, 0.25L, 0.5L))
f = Filaments.init(S, ClosedFilament, N, QuinticSplineMethod())We then perturb each point of the filament with random translations in the
using FFTW: fft, bfft, fftfreq
w_hat = zeros(ComplexF64, N) # perturbation in Fourier space
ks = fftfreq(N, 2π * N / L) # wavenumbers associated to perturbation, in [-N/2, +N/2-1]
kmax = π * N / L
kmax_perturb = 0.5 * kmax # perturb up to this wavenumber (to avoid issues at the discretisation scale)
A_rms = 1e-6 * L # perturbation amplitude (wanted rms value of ws[:])
using StableRNGs: StableRNG # for deterministic random number generation
rng = StableRNG(42) # initialise random number generator (RNG)
for i in eachindex(w_hat, ks)
kabs = abs(ks[i])
if 0 < kabs <= kmax_perturb
w_hat[i] = randn(rng, ComplexF64) # perturb all wavenumbers equally
else
w_hat[i] = 0
end
end
factor = A_rms / sqrt(sum(abs2, w_hat))
w_hat .*= factorNow convert to physical space and update filament points:
ws = bfft(w_hat) # inverse FFT
for i in eachindex(f, ws)
w = ws[i]
f[i] += Vec3(real(w), imag(w), 0)
end
update_coefficients!(f)Finally, create the other vortices by mirror symmetry.
# Helper function which creates a new filament by mirror symmetry with respect to a given plane.
function create_mirror(f::AbstractFilament; xplane = nothing, yplane = nothing, zplane = nothing)
planes = (xplane, yplane, zplane)
nplanes = sum(!isnothing, planes)
nplanes == 1 || error("expected exactly a single symmetry plane")
g = Filaments.similar_filament(f; offset = -end_to_end_offset(f)) # opposite end-to-end offset
for i in eachindex(f, g)
j = lastindex(f) + 1 - i # we switch the orientation of the filament
g[i] = map(f[j], planes) do x, xmid
xmid === nothing ? x : (2 * xmid - x)
end
end
update_coefficients!(g)
g
end
fx = create_mirror(f; xplane = L/2) # mirror symmetry wrt x = L/2 plane
fy = create_mirror(f; yplane = L/2) # mirror symmetry wrt y = L/2 plane
fxy = create_mirror(fx; yplane = L/2) # mirror symmetry of fx wrt y = L/2 plane
fs = [f, fx, fy, fxy]
plot_filaments(fs)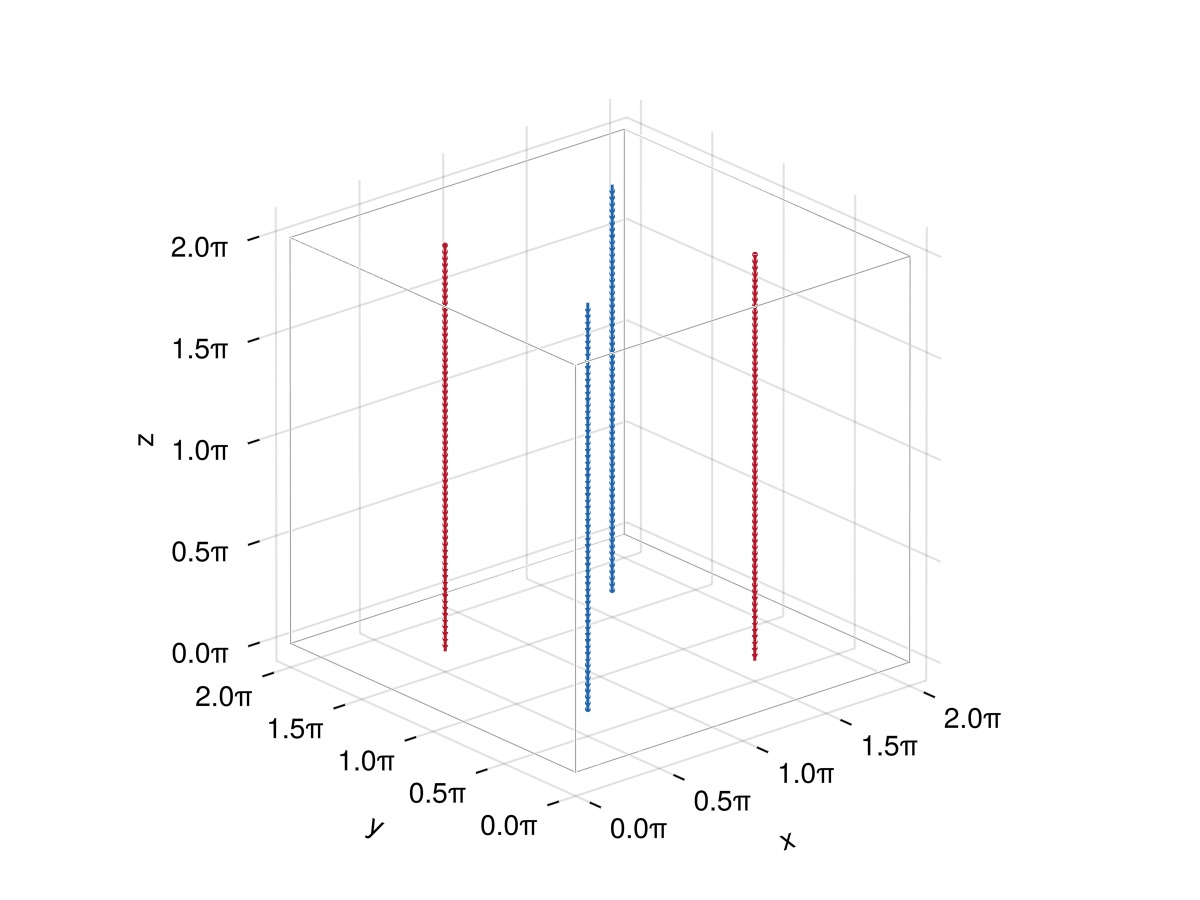
We can now take a look at the wave action spectrum associated to this initial condition to check that, this time, the excited wavenumbers have roughly the same amplitudes:
xs = [s⃗.x for s⃗ in f] # x locations
ys = [s⃗.y for s⃗ in f] # y locations
ws = @. xs + im * ys
w_hat = fft(ws) ./ N # normalised FFT
ks_pos, nk = wave_action_spectrum(ks, w_hat)
nk_normalised = nk ./ A_rms^2
fig = Figure()
ax = Axis(
fig[1, 1];
xscale = log10, yscale = log10, xlabel = L"k", ylabel = L"n(k) / A_{\text{rms}}^2",
xlabelsize = 20, ylabelsize = 20,
xticks = LogTicks(0:4), xminorticksvisible = true, xminorticks = IntervalsBetween(9),
yminorticksvisible = true, yminorticks = exp10.(-40:10),
)
scatterlines!(ax, ks_pos, nk_normalised)
xlims!(ax, 0.8 * ks_pos[begin], nothing)
fig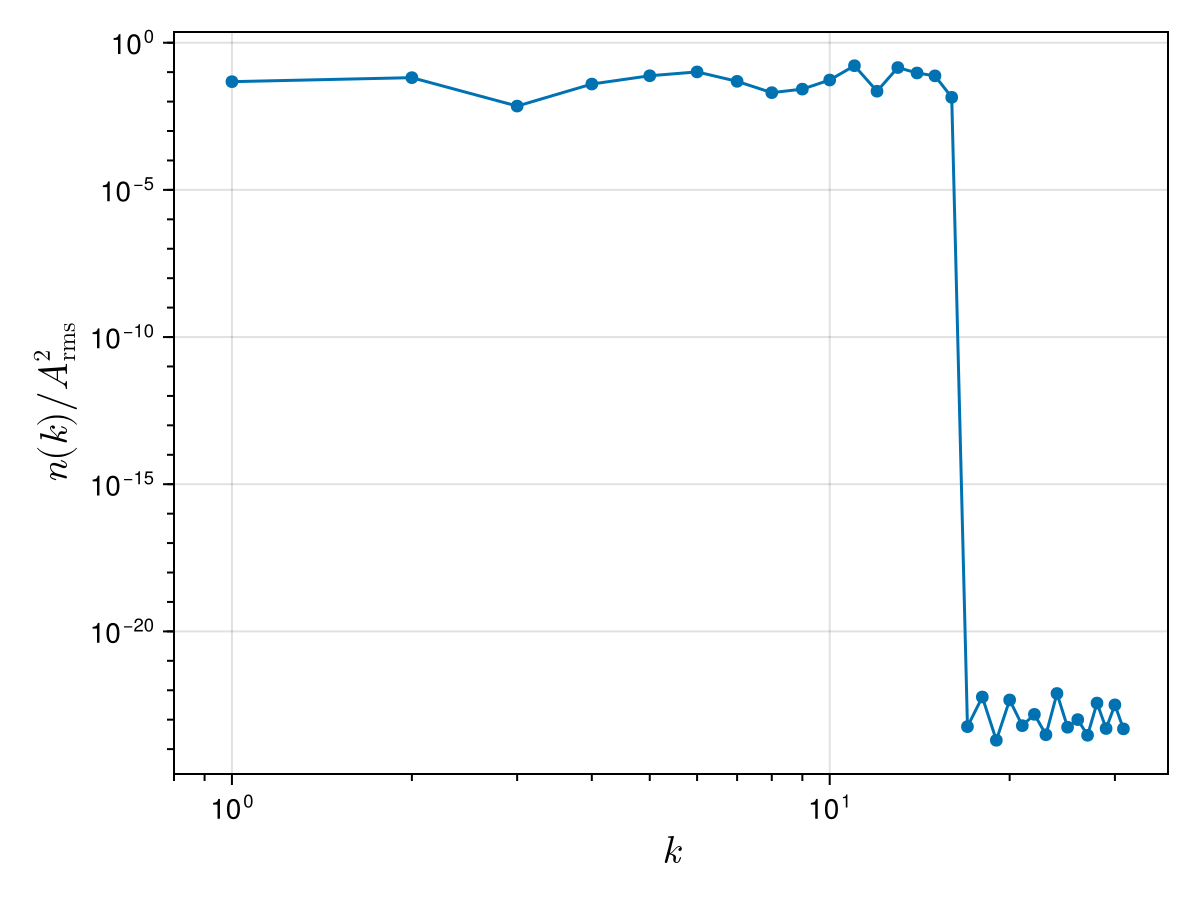
Running the simulation
We create a new problem with this initial condition and with the same parameters as before (except for a longer simulation time):
Tsim = kelvin_wave_period(L; a, Δ, Γ)
prob = VortexFilamentProblem(fs, Tsim, params)VortexFilamentProblem with fields:
├─ p: ParamsBiotSavart{Float64}(Γ = 1.0, a = 1.0e-8, Δ = 0.25, α = 1.4285714285714286, …)
├─ tspan: (0.0, 4.202824549610985) -- simulation timespan
└─ fs: 4-element VectorOfVectors -- vortex filaments at t = 0.0We want to store the whole history of complex positions
const ws_vec = ComplexF64[]
function callback_random(iter)
(; nstep, t,) = iter
if nstep == 0 # make sure vectors are empty at the beginning of the simulation
empty!(times)
empty!(energy)
empty!(ws_vec)
end
E = Diagnostics.kinetic_energy(iter)
push!(times, t)
push!(energy, E)
# Save locations of the first filament in complex form
for s⃗ in iter.fs[1]
w = s⃗.x + im * s⃗.y
push!(ws_vec, w)
end
nothing
end
δ = minimum_node_distance(prob.fs) # should be close to L/N in our case
dt_kw = kelvin_wave_period(δ; a, Δ, Γ)
scheme = Strang(RK4(); nsubsteps = 2)
dt = 2 * scheme.nsubsteps * dt_kw
iter = init(prob, scheme; dt, callback = callback_random, fast_term = LocalTerm())
reset_timer!(iter.to) # to get more accurate timings (removes most of the compilation time)
solve!(iter)
iter.to────────────────────────────────────────────────────────────────────────────────
Time Allocations
─────────────────────── ────────────────────────
Tot / % measured: 20.2s / 94.1% 1.03GiB / 85.4%
Section ncalls time %tot avg alloc %tot avg
────────────────────────────────────────────────────────────────────────────────
Update values at n... 15.2k 16.4s 86.7% 1.08ms 780MiB 86.3% 52.7KiB
Biot-Savart (exc... 1.60k 6.94s 36.6% 4.35ms 232MiB 25.7% 149KiB
Long-range com... 1.60k 4.95s 26.1% 3.10ms 159MiB 17.6% 102KiB
Vorticity to... 1.60k 2.57s 13.5% 1.61ms 71.4MiB 7.9% 45.8KiB
Interpolate ... 1.60k 2.08s 11.0% 1.30ms 57.7MiB 6.4% 37.0KiB
Velocity fie... 1.60k 203ms 1.1% 127μs 5.39MiB 0.6% 3.46KiB
Copy point c... 1.60k 82.2ms 0.4% 51.5μs 21.5MiB 2.4% 13.8KiB
Process poin... 1.60k 1.37ms 0.0% 857ns 0.00B 0.0% 0.00B
Synchronise GPU 1.60k 61.8μs 0.0% 38.7ns 0.00B 0.0% 0.00B
Short-range co... 1.60k 1.56s 8.2% 978μs 46.6MiB 5.2% 29.9KiB
Pair interac... 1.60k 1.21s 6.4% 756μs 10.1MiB 1.1% 6.48KiB
Remove self-... 1.60k 210ms 1.1% 132μs 3.88MiB 0.4% 2.49KiB
Copy point c... 1.60k 93.8ms 0.5% 58.8μs 23.9MiB 2.6% 15.3KiB
Process poin... 1.60k 39.8ms 0.2% 24.9μs 5.85MiB 0.6% 3.75KiB
Synchronise GPU 1.60k 62.9μs 0.0% 39.4ns 0.00B 0.0% 0.00B
Add point charges 1.60k 269ms 1.4% 168μs 6.77MiB 0.7% 4.35KiB
Copy output (d... 3.19k 125ms 0.7% 39.2μs 8.61MiB 1.0% 2.76KiB
Wait for async... 3.19k 3.10ms 0.0% 970ns 200KiB 0.0% 64.0B
CPU-only opera... 1.60k 107μs 0.0% 66.9ns 0.00B 0.0% 0.00B
Biot-Savart (full) 798 4.79s 25.3% 6.01ms 163MiB 18.1% 210KiB
Long-range com... 798 3.62s 19.1% 4.53ms 111MiB 12.3% 143KiB
Interpolate ... 1.60k 2.07s 10.9% 1.30ms 57.7MiB 6.4% 37.0KiB
Vorticity to... 798 1.29s 6.8% 1.62ms 35.7MiB 3.9% 45.8KiB
Velocity fie... 798 113ms 0.6% 141μs 2.72MiB 0.3% 3.49KiB
Streamfuncti... 798 93.5ms 0.5% 117μs 2.69MiB 0.3% 3.46KiB
Copy point c... 798 42.1ms 0.2% 52.8μs 10.7MiB 1.2% 13.8KiB
Process poin... 798 731μs 0.0% 916ns 0.00B 0.0% 0.00B
Synchronise GPU 798 30.6μs 0.0% 38.4ns 0.00B 0.0% 0.00B
Short-range co... 798 912ms 4.8% 1.14ms 35.0MiB 3.9% 44.9KiB
Pair interac... 798 626ms 3.3% 784μs 5.15MiB 0.6% 6.61KiB
Remove self-... 798 106ms 0.6% 133μs 1.99MiB 0.2% 2.55KiB
Copy point c... 1.60k 93.0ms 0.5% 58.3μs 21.5MiB 2.4% 13.8KiB
Local term (... 798 66.4ms 0.4% 83.2μs 1.76MiB 0.2% 2.25KiB
Process poin... 798 14.7ms 0.1% 18.4μs 2.92MiB 0.3% 3.75KiB
Synchronise GPU 798 29.1μs 0.0% 36.4ns 0.00B 0.0% 0.00B
Add point charges 798 152ms 0.8% 191μs 3.39MiB 0.4% 4.36KiB
Copy output (d... 1.60k 87.2ms 0.5% 54.6μs 8.14MiB 0.9% 5.22KiB
Wait for async... 1.60k 1.82ms 0.0% 1.14μs 100KiB 0.0% 64.0B
CPU-only opera... 798 505μs 0.0% 633ns 0.00B 0.0% 0.00B
Biot-Savart (LIA... 12.8k 4.33s 22.8% 339μs 314MiB 34.8% 25.2KiB
Add point charges 12.8k 2.23s 11.8% 175μs 54.3MiB 6.0% 4.36KiB
Short-range co... 12.8k 1.52s 8.0% 119μs 196MiB 21.7% 15.7KiB
Local term (... 12.8k 837ms 4.4% 65.5μs 27.3MiB 3.0% 2.19KiB
Copy point c... 12.8k 640ms 3.4% 50.1μs 153MiB 16.9% 12.3KiB
Synchronise GPU 12.8k 507μs 0.0% 39.7ns 0.00B 0.0% 0.00B
Copy output (d... 12.8k 421ms 2.2% 33.0μs 32.9MiB 3.6% 2.64KiB
Wait for async... 12.8k 17.4ms 0.1% 1.36μs 798KiB 0.1% 64.0B
Advect filaments 15.2k 2.22s 11.7% 147μs 98.8MiB 10.9% 6.67KiB
After advection 798 129ms 0.7% 162μs 3.15MiB 0.3% 4.04KiB
Fold and refine 798 125ms 0.7% 157μs 3.02MiB 0.3% 3.87KiB
Reconnect 798 1.39ms 0.0% 1.74μs 62.3KiB 0.0% 80.0B
Prepare interpolab... 798 98.1ms 0.5% 123μs 17.1MiB 1.9% 21.9KiB
Callback 798 66.2ms 0.3% 82.9μs 4.74MiB 0.5% 6.08KiB
Resize timesteppin... 798 1.39ms 0.0% 1.74μs 0.00B 0.0% 0.00B
────────────────────────────────────────────────────────────────────────────────Once again, kinetic energy is quite well conserved:
std(energy) / mean(energy)2.7857063831074222e-11In fact energy slightly decreases:
(energy[end] - energy[begin]) / energy[begin]-9.51663744373487e-11which can be explained by numerical dissipation at the smallest resolved scales. Notice that the spectrum gets populated at large wavenumbers, which is possibly due to the Kelvin wave cascade mechanism:
ws_mat = reshape(ws_vec, N, :) # reinterpret 1D vector as a 2D matrix: ws_mat[i, j] = w(z_i, t_j)
@views _, nk_start = wave_action_spectrum(ks, fft(ws_mat[:, begin]) ./ N)
@views _, nk_end = wave_action_spectrum(ks, fft(ws_mat[:, end]) ./ N)
fig = Figure()
ax = Axis(
fig[1, 1];
xscale = log10, yscale = log10, xlabel = L"k", ylabel = L"n(k) / A_{\text{rms}}^2",
xlabelsize = 20, ylabelsize = 20,
xticks = LogTicks(0:4), xminorticksvisible = true, xminorticks = IntervalsBetween(9),
yminorticksvisible = true, yminorticks = exp10.(-40:10),
)
scatterlines!(ax, ks_pos, nk_start ./ A_rms^2; label = "Initial time")
scatterlines!(ax, ks_pos, nk_end ./ A_rms^2; label = "Final time")
xlims!(ax, 0.8 * ks_pos[begin], nothing)
axislegend(ax; position = (0, 0))
fig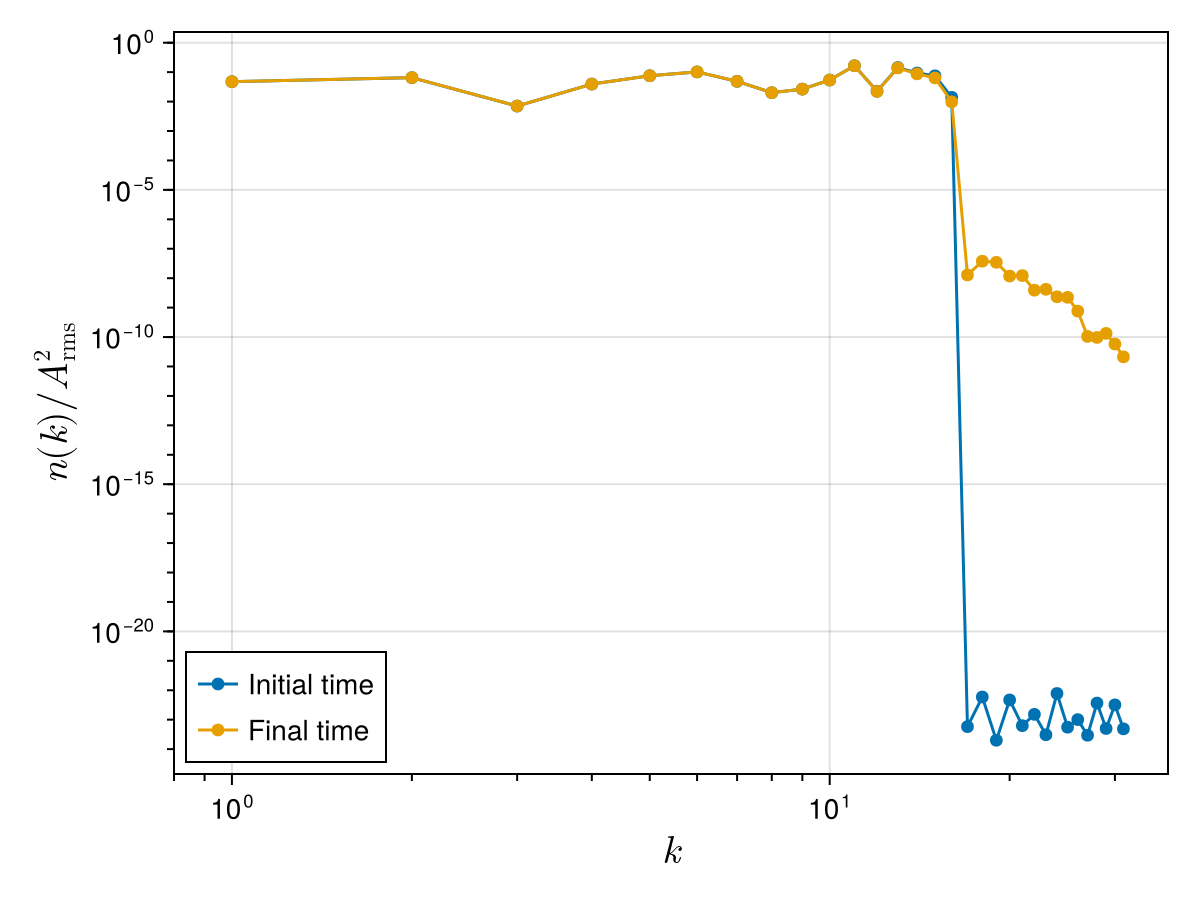
Verifying the Kelvin wave dispersion relation
We will assume that the simulation is in a statistically stationary state, which allows to perform FFTs in time as well as in space.
We start by choosing the times over which we will analyse the data. Currently we analyse all times, but this could be modified in simulations which require some time before reaching a (near-)stationary state.
t_inds = eachindex(times)[begin:end] # this may be modified to use a subset of the simulation time
Nt = length(t_inds) # number of timesteps to analyse
ws_h = ws_mat[:, t_inds] # create copy of positions to avoid modifying the simulation output
ws_h .-= w_equilibrium # subtract equilibrium position64×799 Matrix{ComplexF64}:
-4.15535e-6+4.64122e-6im … -4.2791e-6-1.88914e-6im
1.32043e-6+6.59221e-6im -4.93763e-6+5.3689e-6im
4.9057e-6+7.76414e-6im -4.19102e-6+8.46733e-6im
1.56307e-6+5.96558e-6im -9.88867e-7+3.89358e-6im
-3.99974e-6+2.7909e-7im 2.98489e-6-4.45523e-7im
-3.74592e-6-4.66318e-6im … 3.59489e-6+1.30238e-6im
1.58423e-6-3.24128e-6im -7.28623e-7+4.95071e-6im
3.31877e-6+2.73898e-6im -6.27256e-6+4.86334e-6im
-1.82543e-6+6.06245e-6im -7.55109e-6+1.78905e-6im
-6.59718e-6+4.32956e-6im -2.79821e-6-1.28419e-6im
⋮ ⋱
-1.12477e-5+3.80923e-6im … -9.32702e-6-2.19986e-6im
-3.64297e-6-4.16605e-6im -2.2791e-6+2.61882e-6im
4.25811e-6-7.93425e-6im 3.13886e-6+4.94948e-6im
4.3955e-6-3.3357e-6im 1.52284e-6+1.07325e-6im
5.70624e-7+3.45148e-6im -2.11861e-6-2.59298e-6im
-4.09761e-7+5.23592e-6im … -2.58823e-6+1.51288e-7im
6.03639e-7+2.68405e-6im -1.33013e-6+4.35545e-6im
-1.14974e-6+1.09316e-6im -1.52195e-6+2.15169e-6im
-4.68204e-6+2.39272e-6im -2.99737e-6-3.12112e-6imAs detailed in the Temporal analysis section, we apply a window function over the temporal dimension since the original signal is not exactly periodic in time:
window = DSP.Windows.hanning(Nt)
for i ∈ axes(ws_h, 1)
@. @views ws_h[i, :] *= window
endWe can now perform a 2D FFT and plot the results:
fft!(ws_h) # in-place FFT
ws_h ./= length(ws_h) # normalise FFT
# Combine +k and -k wavenumbers and plot as a function of |k| (results should be symmetric anyways)
ws_abs2 = @views abs2.(ws_h[1:((end + 1)÷2), :]) # from k = 0 to k = +kmax
@views ws_abs2[2:end, :] .+= abs2.(ws_h[end:-1:(end÷2 + 2), :]) # from -kmax to -1
Δt = times[2] - times[1] # timestep
ks_pos = ks[1:(end + 1)÷2] # wavenumbers (≥ 0 only)
ωs = fftfreq(Nt, 2π / Δt) # frequencies
ωs_shift = fftshift(ωs) # for visualisation: make sure ω is in increasing order
w_plot = fftshift(log10.(ws_abs2), (2,)) # FFT shift along second dimension (frequencies)
k_max = ks_pos[end] # maximum wavenumber
ω_max = -ωs_shift[begin] # maximum frequency
# Analytical dispersion relation
ks_fine = range(0, k_max; step = ks_pos[2] / 4)
ωs_kw = @. -Γ * ks_fine^2 / (4 * π) * (
log(2 / (abs(ks_fine) * a)) - γ + 1/2 - Δ
)
# We also plot the wavenumber associated to the mean inter-vortex distance for reference
ℓ = sqrt(params.Ls[1] * params.Ls[2] / length(fs)) # mean inter-vortex distance
k_ℓ = 2π / ℓ
fig = Figure()
ax = Axis(fig[1, 1]; xlabel = L"k", ylabel = L"ω")
xlims!(ax, 0.8 * k_max .* (0, 1))
ylims!(ax, 0.8 * ω_max .* (-1, 1))
hm = heatmap!(
ax, ks_pos, ωs_shift, w_plot;
colormap = Reverse(:deep),
colorrange = round.(Int, extrema(w_plot)) .+ (4, 0),
)
Colorbar(fig[1, 2], hm; label = L"\log \, |\hat{w}|^2", labelsize = 20)
hlines!(ax, 0; color = :white, linestyle = :dash, linewidth = 1)
let color = :lightblue
vlines!(ax, k_ℓ; color, linestyle = :dot)
text!(ax, L"\frac{2π}{ℓ}"; position = (k_ℓ, -0.8 * ω_max), offset = (6, 4), align = (:left, :bottom), color, fontsize = 16)
end
lines!(ax, ks_fine, ωs_kw; color = (:orangered, 1.0), linestyle = :dash, linewidth = 1)
fig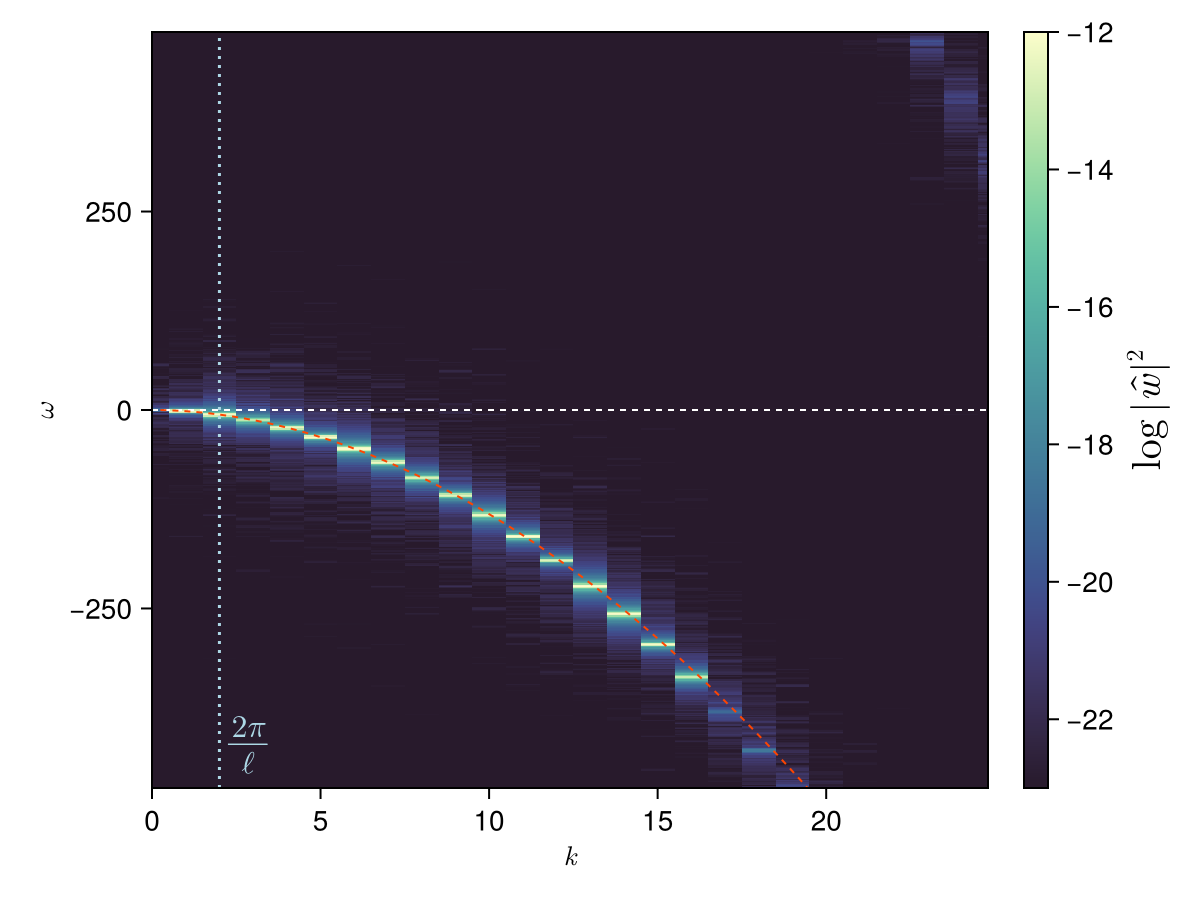
Fluctuations are clearly concentrated on the analytical dispersion relation
This page was generated using Literate.jl.